It was time for a little review of animal cell size control since the last one was 5 years ago from Ginzberg, Kafri and Kirschner (2015). A few things have happened since then including a couple of real molecular models for how cell size might lead to cell cycle progression at the G1/S transition. There is a p38 mechanisms proposed by the Kafri lab that restricts cell cycle progression in small cells, and our own Rb dilution mechanism that is proposed to do the same. Importantly, these mechanisms could operate together to ensure small cells have enough time to grow in G1 before entering the cell cycle. In addition, we discuss the need for better cell culture models that recapitulate the strong G1 size control that Mimi Xie observed takes place in vivo in mouse epidermal stem cells. I hope the review is useful to summarize the last 5 years and to set the stage for what I think will be an explosive next 5 years for our field. Here is a link to our current thoughts.
Read all about it on BioRxiv here https://www.biorxiv.org/content/10.1101/815084v1.full
The abstract says it all:
“The coordination of metabolism and growth with cell division is crucial for proliferation. While it has long been known that cell metabolism regulates the cell division cycle, it is becoming increasingly clear that the cell division cycle also regulates metabolism. In budding yeast, we previously showed that over half of all measured metabolites change concentration through the cell cycle indicating that metabolic fluxes are extensively regulated during cell cycle progression. However, how this regulation is achieved still remains poorly understood. Since both the cell cycle and metabolism are regulated to a large extent by protein phosphorylation, we here decided to measure the phosphoproteome through the budding yeast cell cycle. Specifically, we chose a cell cycle synchronisation strategy that avoids stress and nutrient-related perturbations of metabolism, and we grew the yeast on ethanol minimal medium to force cells to utilize their full biosynthetic repertoire. Using a tandem-mass-tagging approach, we found over 200 sites on metabolic enzymes and transporters to be phospho-regulated. These sites were distributed among many pathways including carbohydrate catabolism, lipid metabolism and amino acid synthesis and therefore likely contribute to changing metabolic fluxes through the cell cycle. Among all one thousand sites whose phosphorylation increases through the cell cycle, the CDK consensus motif and an arginine-directed motif were highly enriched. This arginine-directed R-R-x-S motif is associated with protein-kinase A, which regulates metabolism and promotes growth. Finally, we also found over one thousand sites that are dephosphorylated through the G1/S transition. We speculate that the phosphatase Glc7/ PP1, known to regulate both the cell cycle and carbon metabolism, may play an important role because its regulatory subunits are phospho-regulated in our data. In summary, our results identify extensive cell cycle dependent phosphorylation and dephosphorylation of metabolic enzymes and suggest multiple mechanisms through which the cell division cycle regulates metabolic signalling pathways to temporally coordinate biosynthesis with distinct phases of the cell division cycle.”
It is a very accurate, Jenny-like abstract… I tried to trim, but not easy… Also, many thanks to Josh Elias and Lichao for the nice collaboration on the phosphoproteomics experiments and analysis. Nice work everyone!
Congratulations to mimi on a very nice piece of work.
Here is the link to the bioRxiv paper https://www.biorxiv.org/content/10.1101/754424v1
It is always great when you can recycle existing data. In this case, Mimi analyzed some beautiful data from the Greco lab Mesa et al 2018 link here where they revisit and measure the same epithelial cells on a mouse epithelium for a week. Fantastic data!
tl;dr synopsis: cells exhibit much better size control in vivo than in vitro.
Recent work from many labs has explored size control in animal cells in vitro and concluded that they exhibit an adder, where a constant amount of volume is added in each cell division cycle (Ginzberg, et al., Elife (2018), Cadart, et al., Nat Comm (2018)). However, to date, there are no comparable studies of mammalian cells in vivo so the physiological relevance of adder mechanisms is unclear.
Mimi performed a single cell analysis of cell size control in epidermal stem cells growing and dividing in a living mouse under normal tissue turnover. She quantified the 3D volume growth and cell cycle progression of epidermal stem cells and found that cell growth is coupled to division through a sizer mechanism operating largely in the G1 phase. I.e., regardless of their size at birth, cells transition through G1/S at similar sizes. Thus, while in vitro tissue culture studies identified adder mechanisms, our work demonstrates that sizer mechanisms are important in vivo. Indeed, cells in vivo exhibit a tighter size control than tissue culture cells in vitro.
The last such meeting was 2016 in Berlin and is was a great first meeting for the community. It was fun to see almost 100 people working on my favorite subject. I hope this meeting will even exceed that as we have been helping the field to grow. Looking forward to it!
Read all about it here https://www.sciencedirect.com/science/article/pii/S1097276519302813?dgcid=author
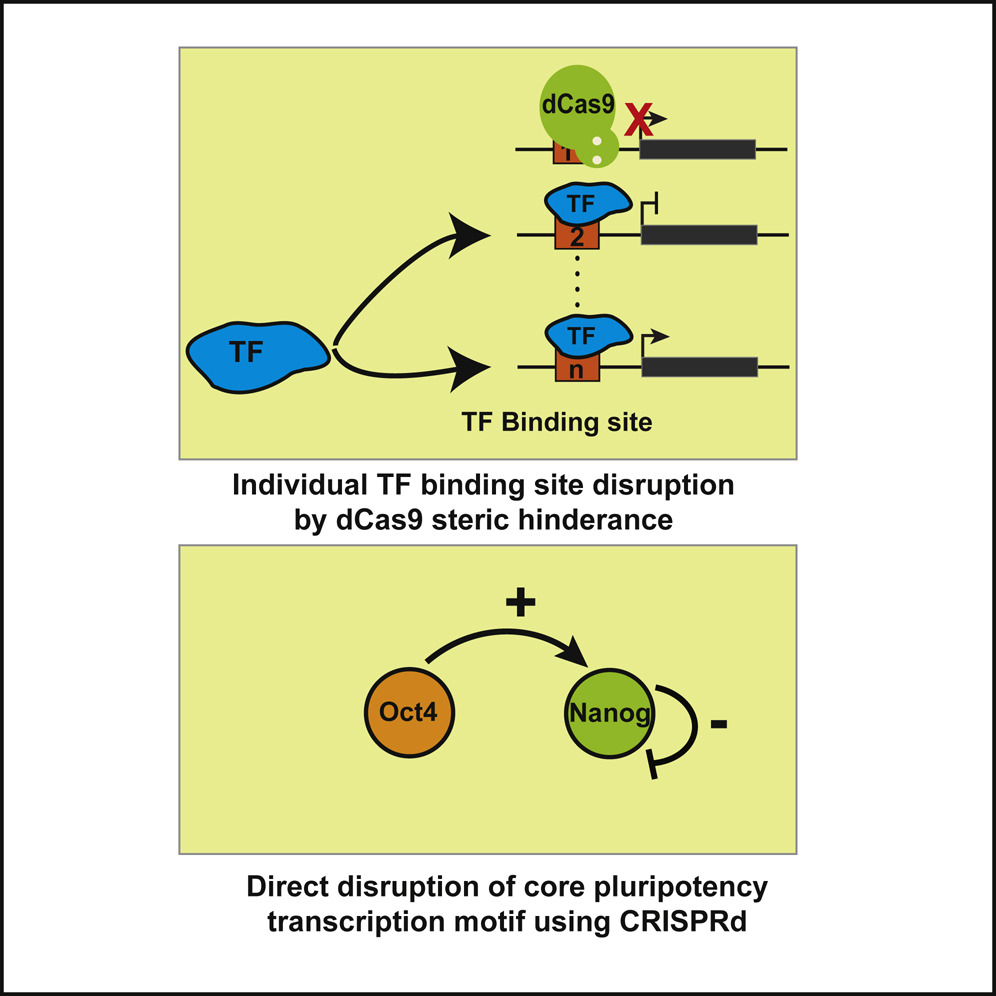
Ali shows that targeting dCas9 to specific transcription factor binding sites can be used to inhibit those sites function. Basically, dCas9 outcompetes the TF to block binding. This worked in all the cases we looked at and resulted in increased and decreased transcription depending on the site blocked. As a case study, we examined the pluripotency network in embryonic stem cells and found that an Oct4 site on the Nanog promoter drove positive feedback to maintain pluripotency, and that a Nanog site in the Nanog promoter had the opposite effect and provided negative feedback. This method can be used to individually delete links in transcriptional networks to asses their function without deleting the transcription factor hubs. It was a nice collaboration with Marius Wernig (stem cell inst) and Stanley Qi and Antonia Dominguez (bioengineering). Thanks everyone for making this such a fun collaboration.